Recent News
Recent Publications
|
Genetically engineered probiotics
My proposed research platform will focus on three areas: 1) developing new genetic tools for cellular communication in prokaryotes and eukaryotes; 2) establishing new hosts for the production of bioactive compounds and 3) unifying my investigations in dynamic regulation and natural product synthesis towards the application of genetically engineered probiotics to treat gastrointestinal diseases.
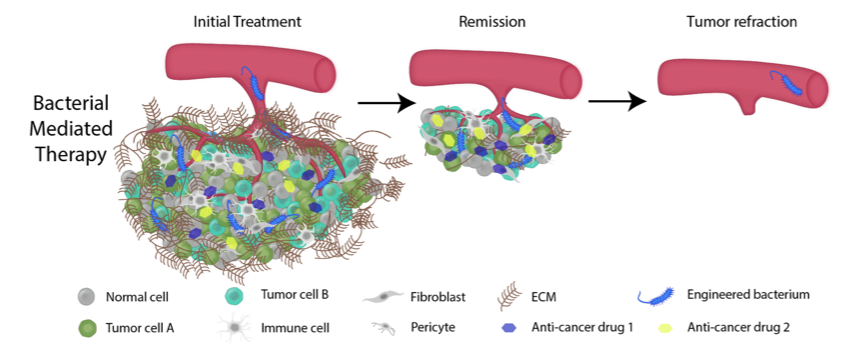
A genetically engineered bacterium can overcome the multifactorial challenges of drug resistance. The bacteria can actively target the tumor site without producing the anticancer compound, reducing off-site toxicity. After reaching the tumor, the bacteria can burrow past the extensive ECM into the hypoxic tumor core. Once the bacteria has colonized the tumor, genetic switches can activate multiple therapies that target parallel disease pathways resulting in complete tumor elimination.
Background
During my PhD, I established a foundation in synthetic biology, a rapidly advancing discipline that impacts many aspects of society. With my background in mathematical modeling during my MS in chemical engineering, I sought to bring engineering rigor to my studies in cellular communication under the guidance of Dr. William Bentley. I elucidated the mechanisms in intra- and interkingdom communication systems and leveraged this understanding to genetically engineer bacteria for bioengineering and biomedical applications.
As a postdoctoral scholar in Dr. Jay Keasling’s laboratory, I have learned to design and manipulate the enzymes and pathways that lead to high-value products. I was awarded the NIH F32 National Research Service Award (NRSA) fellowship to develop an in-depth expertise in polyketide synthases (PKSs), a modular assembly-like enzyme that is responsible for the production of many pharmaceutical drugs. Informed by phylogeny and based in chemoinformatics, my work has advanced a new paradigm in manipulating these enzymatic pathways, and we have applied this knowledge to programmatically produce high-value chemicals in Streptomyces albus. By marrying my graduate school expertise of dynamic regulation with my knowledge in metabolic pathways, I am developing a generalizable platform to produce disease-fighting molecules in the gastrointestinal system. Through a just-in-time release mechanism, genetically engineered probiotics will release therapeutics only at the disease site and only when sufficiently accumulated. I submitted an NIH K99 Transition to Independence application as a therapy to treat colon cancer, which would represent a vertical advance in cancer probiotic therapy. |