|
Our publication in Current Opinion in Biotechnology entitled, "Leveraging microbial biosynthetic pathways for the generation of 'drop-in' biofuels" is now available online. Thank you and congratulations to all my co-authors.
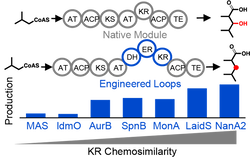
Advances in retooling microorganisms have enabled bioproduction of ‘drop-in’ biofuels, fuels that are compatible with existing spark-ignition, compression-ignition, and gas-turbine engines. As the majority of petroleum consumption in the United States consists of gasoline (47%), diesel fuel and heating oil (21%), and jet fuel (8%), ‘drop-in’ biofuels that replace these petrochemical sources are particularly attractive. In this review, we discuss the application of aldehyde decarbonylases to produce gasoline substitutes from fatty acid products, a recently crystallized reductase that could hydrogenate jet fuel precursors from terpene synthases, and the exquisite control of polyketide synthases to produce biofuels with desired physical properties (e.g., lower freezing points). With our increased understanding of biosynthetic logic of metabolic pathways, we discuss the unique advantages of fatty acid, terpene, and polyketide synthases for the production of bio-based gasoline, diesel and jet fuel.
3 Comments
 Based on the mathematical model we developed in our paper in Metabolic Engineering, we designed a bacterium with the highest rate of AI-2 uptake to manipulate QS behaviors from inside a biocompatible capsule. While in our prior work, we showed that these controller cells could uptake AI-2 when interacting directly with the population, useful in many infections, these cells are retained within the capsule, while AI-2 is small enough to freely diffuse into the capsule and be consumed. By minimally interacting with the cell population, the device could not only quench QS, but could also tune the phenotypes of QS subpopulations. There would be tremendous advantages in controlling the signal intensity so that some cells could make one designed protein, and some cells could make another, thereby generating ‘quantized quorums’, a concept advanced in a previous study (ISME 2016). This was a major leap forward from our work published in the ISME journal, where quantized quorums were generated by knocking out the luxS gene and adding metabolites. Now, we could guide one cell population to produce a product and another subpopulation to produce another, which is the major advantage of microbial consortias. Following up on this work, I described how discrete levels of molecules like AI-2 could be used to connect different cell populations together on a microfabricated chip as a ‘bioproduction breadboard’ (cover, Curr. Opin. 2016). By separating the populations but linking and coordinating them with discrete levels of AI-2, we could create microbial communities with balanced growth populations. This allows de novo biological pathways to be produced by leveraging the specialization of each cell type. I am happy to announce that my paper entitled "Enhancing Intercellular Coordination: Rewiring Quorum Sensing Networks for Increased Protein Expression through Autonomous Induction" is now available online. In this paper, my co-authors and I create a genetically engineered bacterium with advanced quorum sensing kinetics. These cells produce their own coordination molecules and then rapidly uptake these molecules, which trigger protein expression. This enhanced uptake increases the coordination among the bacteria, resulting in increased protein expression. As induction molecules for protein expression are cost-prohibitive at industrial scale, we envision these kinds of systems will prove beneficial in metabolic engineering applications.
My first-author publication, "A 'bioproduction breadboard': programming, assembling and actuating cellular networks" is now available online! In this paper, we advance the concept of microfabricated bioproduction device that links diverse cellular populations for the creation of de novo pathway design.
ISME has published our paper entitled "Directed assembly of a bacterial quorum." This paper advances the concept of controlling population phenotypes through quorum sensing. Congratulations to all the authors!
I am happy to announce that my first author paper in Metabolic Engineering has been published. In this work, we developed 'controller cells' that modulate the quorum sensing signal, AI-2, and applied these cells to silence synthetic QS networks that produce recombinant proteins and manipulate natural QS networks that contribute to biofilm formation and chemotaxis. Congratulations to all my co-authors and collaborators. The publication can be found here
I was happy to give a talk at the national meeting of ACS in Denver entitled ‘Controller cells’ that guide communication and enable tunable gene expression
I am very excited to announce that my first-author paper entitled "Bacterial Secretions of Nonpathogenic Escherichia coli Elicit Inflammatory Pathways: a Closer Investigation of Interkingdom Signaling" has been accepted for publication to mBio. In this paper, we perform a global transcriptome study of the effects of the bacterial 'secretome' (secreted products) on epithelial cells, revealing several activated signaling pathways. Further exploration will aid in the understanding of microbial disease, and modulation of existing interkingdom signaling networks may result in novel methods to combat infections. Thank you and congratulations to my fellow co-authors and collaborators.
|
Amin ZargarBS Chemical Engineering Archives
June 2017
|
